nematodes are a model system to study development
Nematodes (roundworms) are simple animals. Adults of the best-studied species, Caenorhabditis elegans, shown on the right, are about 1mm long. They go through two generations per week and have a very rapid embryonic development. They are amenable to forward and reverse genetics approaches, and a community of hundreds of researchers studies them. |

our focus: gene networks in development
During embryonic development, cells must choose from among different types of tissues to make through the process of Cell Fate Specification. In nematodes like C. elegans, some of these decisions are made very early in development, within hours after fertilization. When specification does not occur properly, the embryo will be missing one or more tissues. We have found that the genes that direct specification of two lineages -- those producing endoderm and mesoderm -- form a complex transcriptional gene regulatory network. The same process of specification occurs in all animals, and in adult stem cells, for example in the hematopoietic lineages that produce blood and immune cells.

the C. elegans intestine is derived from the E cell
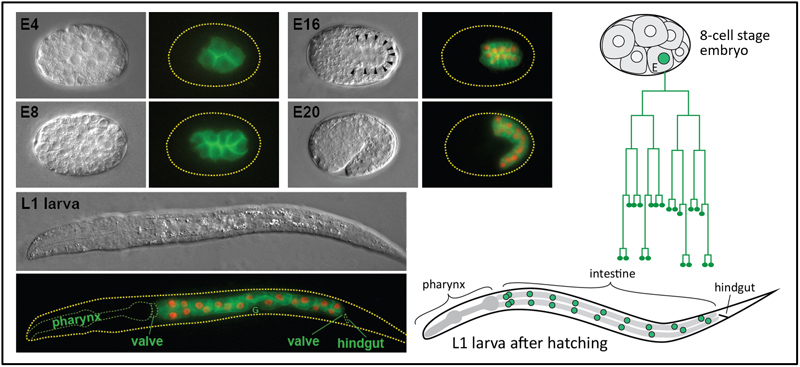
The C. elegans intestine has 20 cells that are descendants of the E cell, found at the 8-cell stage (top right). Micrographs on the left show the developing gut at various stages of embryogenesis. A GFP marker indicates the intestinal cell membranes, and red in later stages shows the intestinal nuclei.
→ more about intestine development

a gene regulatory network specifies the intestinal fate
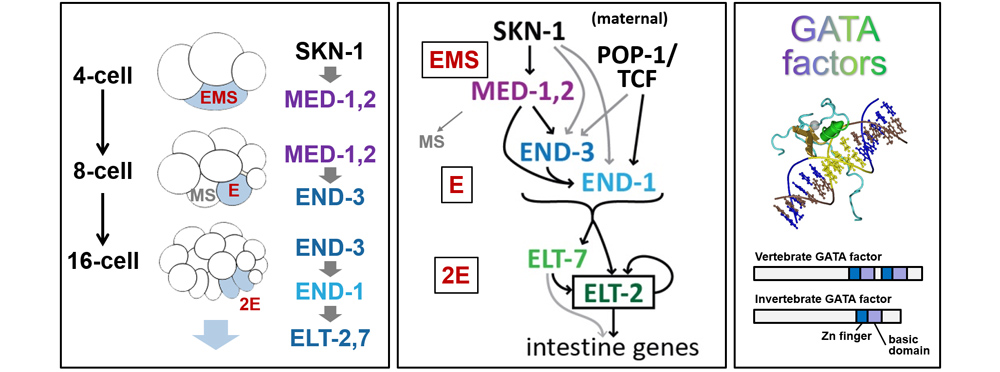
Work by our group and others has shown that in C. elegans, a cascade of transcription factors is expressed in the early E lineage that directs the development of the intestine. Most of these (in purple, blue and green) belong to the GATA factor family. The multiple GATA factors exhibit a fair bit of redundancy: Mutation of any single factor has a weak phenotype, loss of end-1 and end-3 together results in a complete failure to specify the gut, and other combinations result in stochastic gut specification. In other work, we elucidated how the genes tbx-35 and ceh-51 work downstream of med-1,2 to specify the mesodermal fate of MS, the sister cell of E.
→ gut specification → stochasticity → mesoderm specification

evolution of gut specification in Caenorhabditis
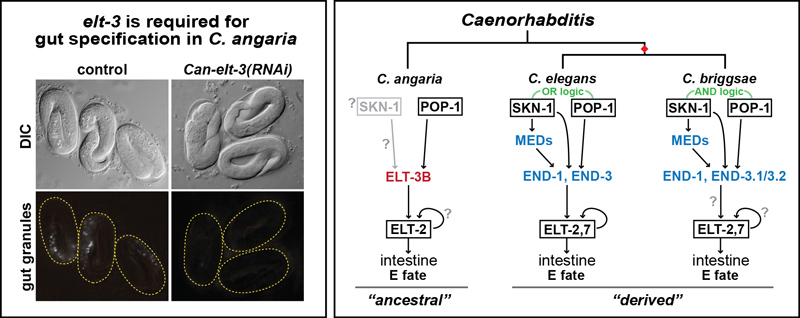
We have recently studied evolution of the endodermal GATA factors across the genus, and found that the MED and END factors (and ELT-7) do not exist outside of the Elegans Supergroup of species closely related to C. elegans. We found that the ELT-3 factor, which exists in C. elegans but does not have a role in gut specification, is the likely ancestral gut specification factor outside of the Elegans Supergroup. Above left: In the species C. angaria, knockdown of elt-3 by RNA interference results in the loss of gut, analogous to loss of end-1,3 in C. elegans. Above right: We hypothesize that a simpler network for gut specification, as seen in C. angaria, underwent an expansion at the base of the Elegans Supergroup (red diamond) to form the more complex network seen in C. elegans and C. briggsae. The rewiring of the gene network is an example of Developmental System Drift.
→ evolution of gut factors → C. briggsae → C. angaria → DSD in gut specification

gut development in Pristionchus
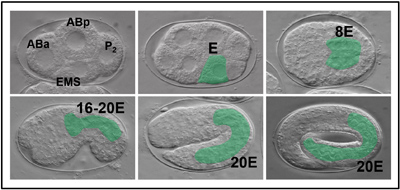 |
Nematodes of the genus Pristionchus are used as a comparison species to C. elegans. On the left are embryonic stages of P. pacificus development showing the cells at the 4-cell stage and the development of the E lineage (shaded green). We are characterizing the GRN that specifies gut in P. pacificus and related species. So far, we have found evidence for a similar expansion of an ancestral gut specification network within the genus, similar to what is hypothesized to have occurred in Caenorhabditis. |
