
 |
Morris F. Maduro Div. Dean of Life Sciences, CNAS Prof. of Biology, Dept. of MCSB More info here. |
 |
Gina Broitman- Maduro Research Associate |
 |
Kortney Schoenborn RISE 2024 |
 |
Allison Matthews RISE 2024 |
 |
Dylan Truong RISE 2024 |
 |
Akash Verma Biology Major |
 |
Ryan Dang RISE 2025 |
 |
Preston Ambrose RISE 2025 |
 |
Melissa Guthrie PhD Student (CMDB Program) |
 |
Charles Warden Rotating GGB Student |
Structure and Evolution of Gene Regulatory Networks
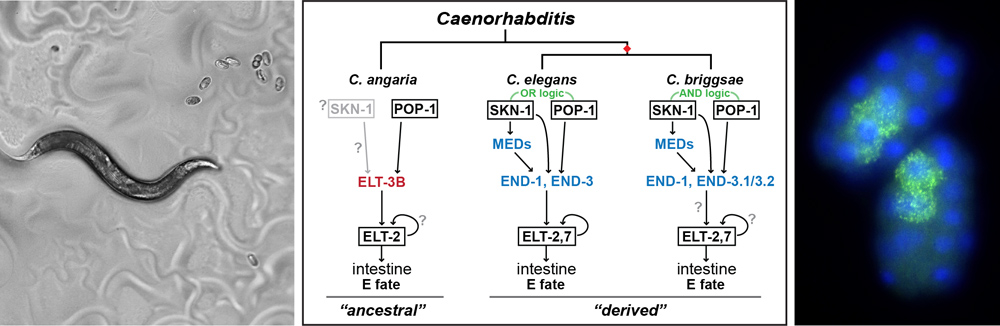
Transcriptional Gene Regulatory Networks (GRNs) direct the development of multicellular animals, and are important in other contexts, including stem cell specification and development of cancer. Developmental GRNs typically exhibit layers of redundancy to assure the robust specification of early embryonic precursor cells. Changes in such networks are generally known to underlie differences in form and function between related species. Paradoxically, GRNs can also undergo evolutionary changes even when there is no apparent change in development, a phenomenon called Developmental System Drift. We have studied the specification of the embryonic endoderm precursor cell E in the nematode C. elegans and its close relatives for more than 20 years. In more distantly related species, the gene network for specifying the endoderm is simpler and has fewer factors, suggesting that the more complex network in C. elegans was derived from a simpler ancestral network. We are currently extending this work into nematodes of the genus Pristionchus. Our work promises to yield new insights into how gene networks evolve over time.
→ More information about our research here.
News Highlights
| — July 1, 2025 - Worm Show at the International Worm Meeting! — |
Curtis Loer and MM presented the ninth Worm Show at UC Davis More information is here. |

| — June 17-20, 2024 - EvoWorm Meeting in Vienna — |
 2024 Worm Evolution Meeting at the Vienna BioCenter in Vienna, Austria. Clockwise from upper left: Group image; MM with Ron Ellis; Gina at our poster; Gina with Oliver Hobert and André Pires da Silva; meeting organizers Yen-Ping Hsueh, Amanda Gibson, Gavin Woodruff, Erik Andersen, and Alejandro Burga |

| — April 8, 2024 - Total Solar Eclipse! — |
 We watched the Great American Eclipse of 2024 from Franklin, Indiana, where totality was just over four minutes. Read another article about MM's eclipse hobby and the 2024 trip, here. (Canon R6mkII, 800mm f/11) |

| — January, 2024 - NSF Grant Awarded! — | |
 |
We have been awarded a $1.3M grant from the National Science Foundation to study developmental system drift in endoderm specification, using Pristionchus. |

| — April 5, 2023 - Our paper on C. angaria was a finalist for Outstanding Paper — | |
 |
Our 2022 paper on gut specification in C. angaria, a distant relative of C. elegans, was one of three finalists for from the journal Development. |
→ See all news here.
Recent Papers
Broitman-Maduro G, Maduro M (2023). Evolutionary change in gut specification in Caenorhabditis centers on the GATA factor ELT-3 in an example of Developmental System Drift. J. Dev. Biol. 11, 32. https://doi.org/10.3390/jdb11030032 ![]()
![]()
Broitman-Maduro G, Maduro M (2023). The long isoform of the C. elegans ELT-3 GATA factor can specify endoderm when overexpressed. microPublication Biology. https://doi.org/10.17912/micropub.biology.000748 ![]()
![]()
Broitman-Maduro G, Sun S, Kikuchi T, Maduro M (2022). The ELT-3 GATA factor specifies endoderm in Caenorhabditis angaria in an ancestral gene network. Development (2022) 149 (21): dev200984. https://doi.org/10.1242/dev.200984
• Commented on in a Research Highlight. • UCR Press release here. • Finalist for Outstanding Paper of 2022 from Development (link). ![]()
![]()
→ See all papers here.
