Research Focus
Project Description:
Malaria is caused by a protozoan Apicomplexan parasite, Plasmodium. Responsible for 1-3 million deaths per year, malaria is one of the most deadly infectious diseases in the world.
Today, the social and economic impact at both individual and governmental levels, the absence of a vaccine and the spread of parasite resistance to commonly use and inexpensive antimalarial drugs hastens the need for new therapeutic approaches.
One of the factors limiting the successful development of new antimalarials is our restricted understanding of the parasite’s complex biology. In recent years, sequencing of entire genomes of several Plasmodium species (1-4) marked a significant breakthrough in the fight against the disease and allows a profound investigation of the parasite biology and its complex parasite life cycle’s regulation.
DNA micro-array and mass spectrometry technologies are now being used to systematically characterize transcript and protein patterns across the malaria life cycle and provide important clues regarding the regulation of developmental events and their biological significance (5-7). However, the mechanisms underlying this regulation are still poorly understood and appear to be unconventional compared to those of most model organisms.
Latest observations suggest that classical transcriptional regulation mechanisms do not occur in this unusual organism and accentuate the need for a comprehensive understanding of the complex regulatory networks driving the life cycle progression at the protein level. One of the first steps to fulfill this task is to identify and investigate both P. falciparum proteome-wide interactions as well as post-transcriptional and post-translational modifications during key life cycle stages.
Our primary goal is to focus on one of the main post-translational regulatory elements driving the parasite life cycle progression: The Ubiquitinproteasome pathway.
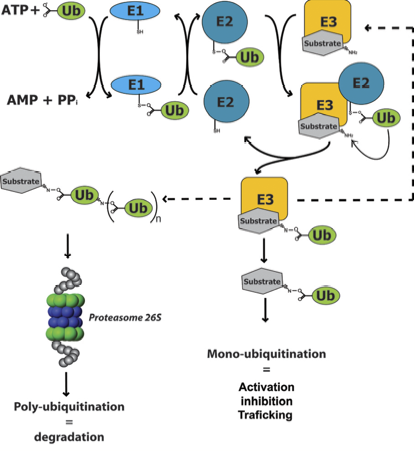
Ubiquitination is one of the major post-translational regulatory events in eukaryotic cells. In particular, the ubiquitin/proteasome system (UPS) serves as the most important protein degradation pathway in eukaryotes. The system involves the labeling of target proteins with a small regulatory protein, ubiquitin, via a cascade of 3 enzymes termed E1, E2 and E3. The ubiquitin-activating enzyme E1 activates the ubiquitin in an ATP dependent manner and transfers it via a thioester intermediate to an ubiquitin-conjugating enzyme, E2. Activated E2 acts with an ubiquitin-ligase, E3, to transfer the ubiquitin to the target substrate, forming an isopeptide bond (Fig 1). Indeed, the UPS temporally controls the proteolysis of key cellular factors involved in processes such as cell cycle, differentiation or development. Beyond the role of protein ubiquitination in proteolysis mediated by the proteasome, it can also regulate protein activation, inhibition, interaction, localization, trafficking, DNA repair and transcription, in a proteasome-independent manner. The importance of the UPS has been widely illustrated by the fact that protein abnormalities in this pathway are implicated in the pathogenesis of many human diseases, including cancer or neurodegenerative diseases such as Alzheimer or Parkinson (12).
Until now, the ubiquitin/proteasome pathway has been largely overlooked in parasite biology. However, our preliminary data have identified more than one hundred UPS components in the genome of several apicomplexan parasites. The UPS in apicomplexan parasites is anticipated to be important for the for two major reasons:
- Considering its conserved role among eukaryotes, this system is expected to regulate key molecular events driving the parasite life cycle, including parasite discrete apicomplexan mechanisms such as host cell invasion and apicoplast formation;
- Components identified as key regulatory events in the infection cycle will be critical for new drug development.
Our fist objective is to identify components of the UPS in apicomplexan parasites.
The apicomplexa phylum comprises a large group of exclusively parasitic organisms, which include a number of significant human pathogens. Complete genome sequences of apicomplexan parasites have recently become available. An examination of these genomes reveals that they have been poorly annotated with more than 60 % of proteins described as “hypothetical”. The main objective of our work is to identify all components of the ubiquitin system using computational analysis. A comprehensive comparative proteomic analysis using the hidden Markov model-base algorithm and additional computational tolls will assist us in the identification of conserved and unique apicomplexan UPS proteins.
Our second objective is to analyze the function of the specific components of the pathway in the context of malaria biology.
Preliminary computational analysis identified 48 ubiquitin ligases in the plasmodium genome. These ubiquitin ligases represent the specific component of the pathway. Most of these enzymes are expressed at particular life cycle stages and are expected to have different functions throughout the life cycle progression (3, 5, 8). Our aim is to understand their exact function using a set of genetic tools to characterize which proteins they interact with throughout the parasite life cycle. To accomplish this, a proteomic approach will be undertaken to identify substrates of the various E3's. Tag-E3 protein will be isolated with their binding partners which will be identify by mass spectrometry. This information should provide insights into molecular events driving the different steps of the life cycle progression. We will mostly focus our research on two specific parasite pathways: The invasion pathway and the transcriptional regulation pathway by chromatin modification.
Role of ubiquitin ligases E3 in invasion
In contrast to other pathogens, apicoplasts can quickly and efficiently enter the cell using their own invasion machinery. This invasion pathway represents a parasite specific process particularly relevant for the parasite virulence. The mechanism of the parasite invasion into its host cell is not yet understood. Several steps and a large collection of proteins have been shown to be involved in this process. However, there is little understanding in the signaling activation of this pathway. Several Plasmodium ubiquitin ligases are co-expressed with proteins invasion such as rhoptry proteins and are expected to have a role in signaling, trafficking, plasticity, endocytosis or exocytosis of these invasion proteins. Our objective is to demonstrate the possible role of the ubiquitin system in the parasite signaling invasion pathway and to provide new insights into molecular events driving this apicomplexan specific process.
Chromatin structure and chromatin modifications
Mechanisms controlling gene expression in the parasite are still poorly understood. Rising evidence indicates that control of gene expression in P. falciparum occurs at multiple levels, one of those being chromatin remodeling. It is increasingly apparent that histone posttranslational modifications are important in chromatin structure. Physiological observations in other organisms suggest that ubiquitinated histones may have multiple functions and structural effects. Determination of chromatin's structural changes resulting from histone ubiquitination is therefore important for the understanding of transcriptional regulation within P. falciparum. We are investigating ubiquitination of histones along the erythrocytic cycle and why these proteins are ubiquitinilated (Degradation/Activation).
Our third and last objective is to validate selected ubiquitin ligase as potential drug targets, and to develop a drug-screening assay using a collection of natural products.
In addition to the fundamental scientific approach, we are validating Ubiquitin ligase as potential drug targets. A knockout strategy is being developed in my lab to validate the importance of this UPS component throughout the parasite life cycle. In parallel, we are developing a drug-screening assay to identify new small molecule inhibitors. An ongoing collaboration with the Scripps Oceanography Institute (San Diego, CA) is providing us with a comprehensive array of marine extracts. Pilot tests laboratory have proved that the process have uncovered compounds that can inhibit malaria growth.
Concluding remarks
With the combined efforts of target identification and small molecule and natural compound screens, we are undividedly searching for a way to break the cycle of malaria infection and resistance.